[1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import sys
import os
import shutil
import warnings
import matplotlib.patches as mpatches
from IPython.display import Image
import pygeostat as gs
%load_ext autoreload
%autoreload 2
%pylab inline --no-import-all
warnings.filterwarnings('ignore')
Populating the interactive namespace from numpy and matplotlib
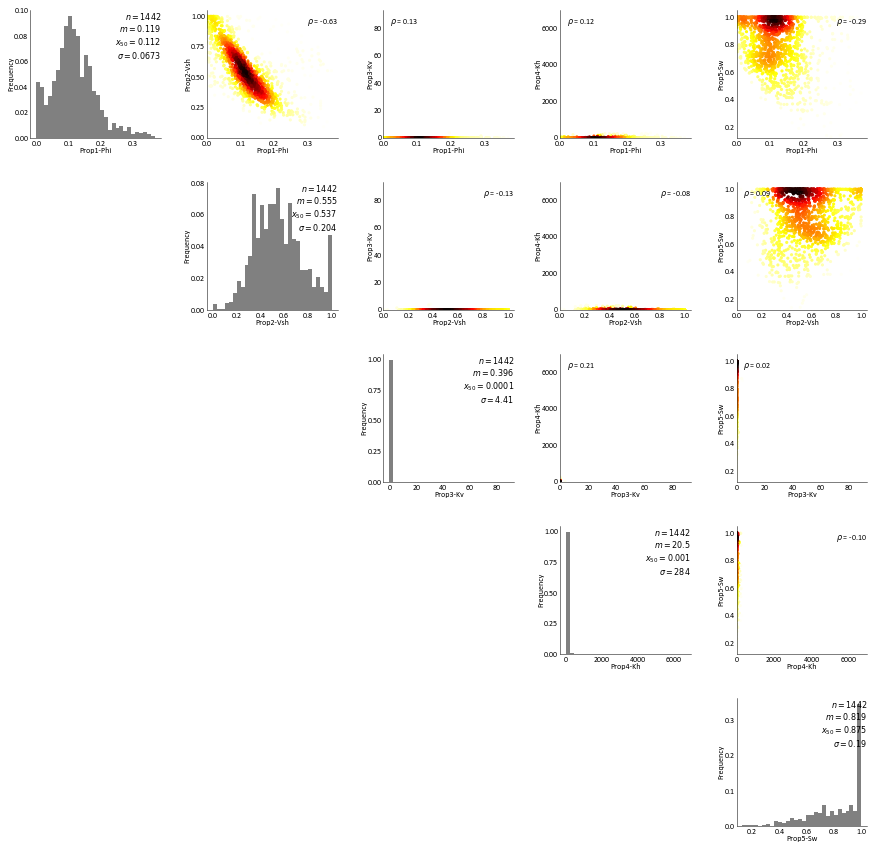
Summary Statistics for Well Data¶
The main statistics for sampled well data are checked with proper visualizations
vtk visulization for a petroleum reservoir model and sampled wells¶
[2]:
Image(filename='data/F1.png')
[2]:
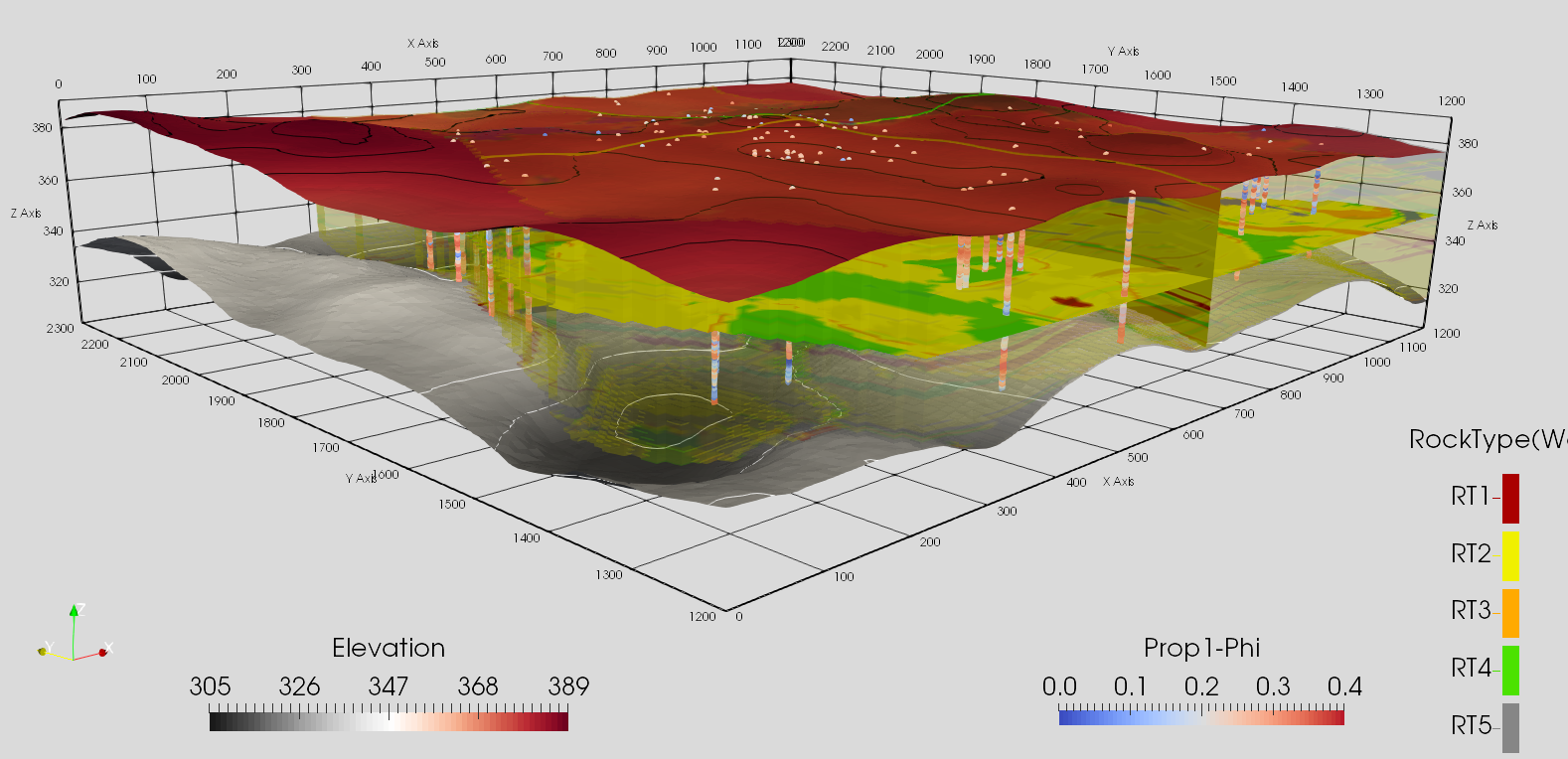
[3]:
Image(filename='data/F2.png')
[3]:
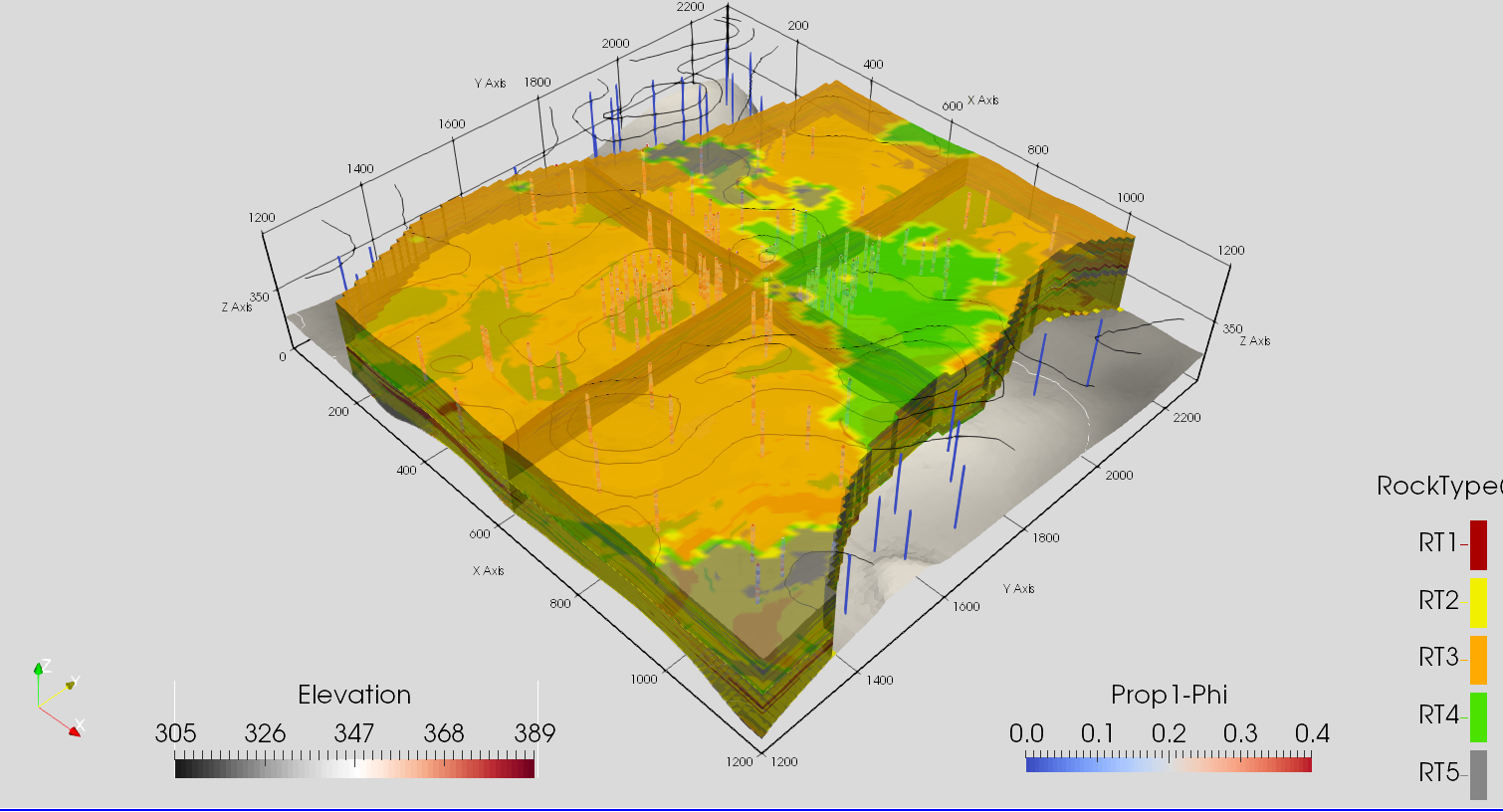
[4]:
Image(filename='data/F3.png')
[4]:
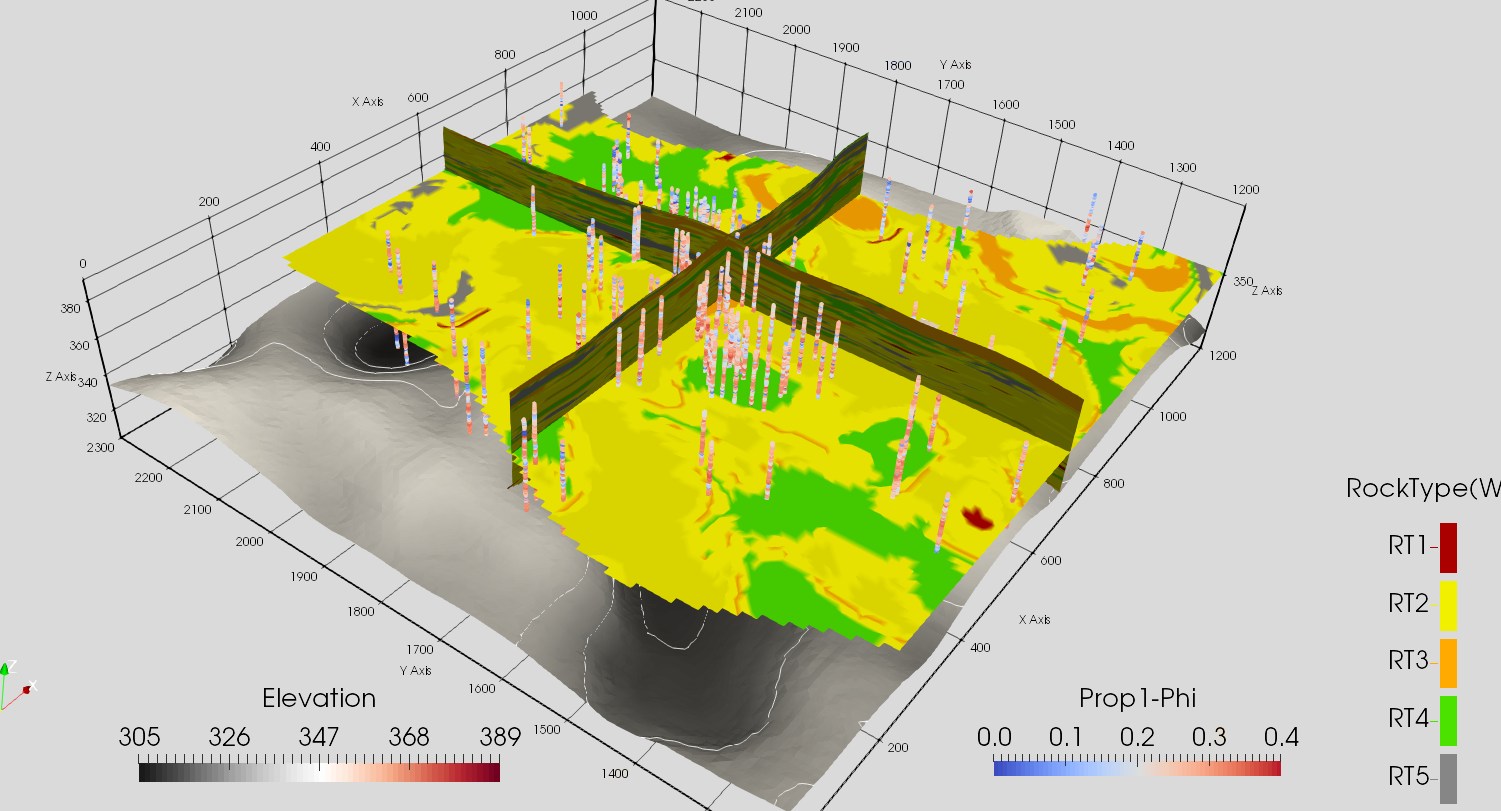
Boundary modelling Data¶
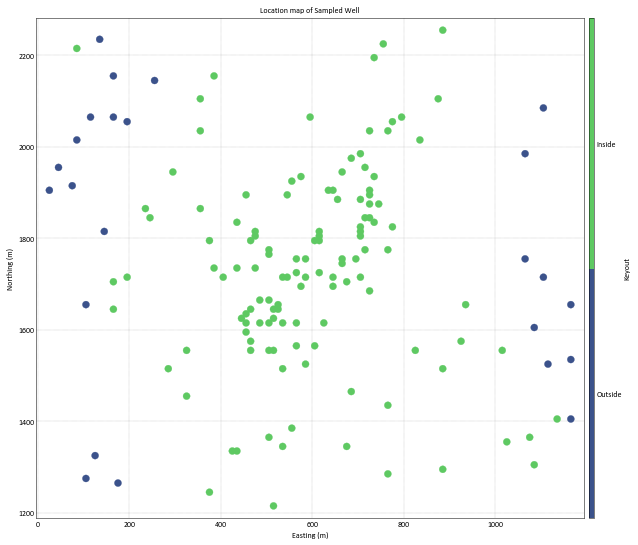
Checking data with keyout (inside/outside) reservoir data. This data will be used for boundary modeling.
[7]:
Boundary_DF = gs.DataFile('data/Baoundary_Final.dat')
Boundary_DF.data.head()
[7]:
| HoleID | X | Y | Keyout | |
|---|---|---|---|---|
| 0 | 1.0 | 85.0 | 2215.0 | 0.0 |
| 1 | 2.0 | 1105.0 | 2085.0 | 0.0 |
| 2 | 3.0 | 405.0 | 1715.0 | 1.0 |
| 3 | 4.0 | 195.0 | 2055.0 | 0.0 |
| 4 | 5.0 | 235.0 | 1865.0 | 1.0 |
[8]:
gs.locmap(Boundary_DF, x='X', y='Y',var='Keyout', figsize=(10,10), cbar_label='Keyout', grid=True,
s=50, catdata=True, catdict={'Outside': 0, 'Inside': 1}, title='Location map of Sampled Well')
[8]:
<mpl_toolkits.axes_grid1.axes_divider.LocatableAxes at 0x18002b99278>
Rock type and Petrophysical Properties¶
[11]:
Well_DF = gs.DataFile('data/SampledWell_Final.dat')
Well_DF.data.head()
[11]:
| HoleID | X | Y | Elevation | Z_Top | Z_Bottom | RockType | Prop1-Phi | Prop2-Vsh | Prop3-Kv | Prop4-Kh | Prop5-Sw | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3.0 | 405.0 | 1715.0 | 330.54334 | 382.79334 | 330.18146 | 5.0 | 0.138597 | 0.421818 | 0.00010 | 0.00100 | 0.783311 |
| 1 | 3.0 | 405.0 | 1715.0 | 331.04334 | 382.79334 | 330.18146 | 5.0 | 0.078082 | 0.666037 | 0.00010 | 0.00100 | 0.922857 |
| 2 | 3.0 | 405.0 | 1715.0 | 331.54334 | 382.79334 | 330.18146 | 3.0 | 0.317862 | 0.215853 | 727.41432 | 1819.14960 | 0.131871 |
| 3 | 3.0 | 405.0 | 1715.0 | 332.04334 | 382.79334 | 330.18146 | 3.0 | 0.397035 | 0.035916 | 8158.62840 | 9486.77730 | 0.118336 |
| 4 | 3.0 | 405.0 | 1715.0 | 332.54334 | 382.79334 | 330.18146 | 1.0 | 0.241755 | 0.235848 | 204.91616 | 697.51595 | 0.475459 |
Get the global proportions for rock types¶
[12]:
RT_code = np.unique(Well_DF.data['RockType'])
RT_name = ['RT%i'%(i) for i in RT_code]
RD_dict = dict(zip(RT_code, RT_name))
RD_dict
[12]:
{1.0: 'RT1', 2.0: 'RT2', 3.0: 'RT3', 4.0: 'RT4', 5.0: 'RT5'}
[13]:
Well_DF.data['RockTypeName'] = Well_DF.data['RockType'].apply(lambda x: RD_dict[x])
Well_DF.data.head()
[13]:
| HoleID | X | Y | Elevation | Z_Top | Z_Bottom | RockType | Prop1-Phi | Prop2-Vsh | Prop3-Kv | Prop4-Kh | Prop5-Sw | RockTypeName | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3.0 | 405.0 | 1715.0 | 330.54334 | 382.79334 | 330.18146 | 5.0 | 0.138597 | 0.421818 | 0.00010 | 0.00100 | 0.783311 | RT5 |
| 1 | 3.0 | 405.0 | 1715.0 | 331.04334 | 382.79334 | 330.18146 | 5.0 | 0.078082 | 0.666037 | 0.00010 | 0.00100 | 0.922857 | RT5 |
| 2 | 3.0 | 405.0 | 1715.0 | 331.54334 | 382.79334 | 330.18146 | 3.0 | 0.317862 | 0.215853 | 727.41432 | 1819.14960 | 0.131871 | RT3 |
| 3 | 3.0 | 405.0 | 1715.0 | 332.04334 | 382.79334 | 330.18146 | 3.0 | 0.397035 | 0.035916 | 8158.62840 | 9486.77730 | 0.118336 | RT3 |
| 4 | 3.0 | 405.0 | 1715.0 | 332.54334 | 382.79334 | 330.18146 | 1.0 | 0.241755 | 0.235848 | 204.91616 | 697.51595 | 0.475459 | RT1 |
[14]:
Proportion = Well_DF.data.groupby(['RockTypeName'],as_index=False).count()
Proportion = Proportion[['RockTypeName', 'RockType']]
Proportion.columns=['RockTypeName', 'Proportion']
Proportion['Proportion'] = Proportion['Proportion'].apply(lambda x: x/len(Well_DF.data))
Proportion
[14]:
| RockTypeName | Proportion | |
|---|---|---|
| 0 | RT1 | 0.042461 |
| 1 | RT2 | 0.392264 |
| 2 | RT3 | 0.233073 |
| 3 | RT4 | 0.220875 |
| 4 | RT5 | 0.111326 |
Uni/Bi variate relationships for different properties within each rock type¶
[15]:
variables = ['Prop1-Phi', 'Prop2-Vsh', 'Prop3-Kv', 'Prop4-Kh', 'Prop5-Sw']
utrivars = [(vx, vy) for j, vx in enumerate(variables) for i, vy in enumerate(variables) if i > j ]
RockType 1¶
[16]:
dat = Well_DF.data[Well_DF.data['RockType']==1]
# setup some custom styles for the output
pltstyle = 'ccgpaper'
cust_style = None
# Upper Triangle: KDE plot
def iter_upper(axes):
for ax, (varx, vary) in zip(axes, utrivars):
gs.kdeplot(dat[varx], dat[vary], ax=ax, density=False, rotateticks=(False, False), cmap="hot_r",
cbar=False, contour=True, lw=0.5, pltstyle=pltstyle, cust_style=cust_style, s=10,
threshold=0.1, shade=False)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Diagonal: Histograms
def iter_diagonal(axes):
for ax, var in zip(axes, variables):
gs.histplt(dat[var], ax=ax, color="grey", pltstyle=pltstyle, cust_style=cust_style,
xlabel=var)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Setup the imagegrid using symmetric, the three functions, and the `unequal_aspects` keyword
# which allows the histograms to be plotted along side the bivariate plots without strange aspect
# issues
fig = gs.imagegrid(upperfunc=iter_upper, diagfunc=iter_diagonal, nvar=len(variables),
symmetric=True, unequal_aspects=True, axes_pad=(0.35, 0.35),
ntickbins=(6,5), pltstyle=pltstyle, cust_style=cust_style, figsize=(15, 15))
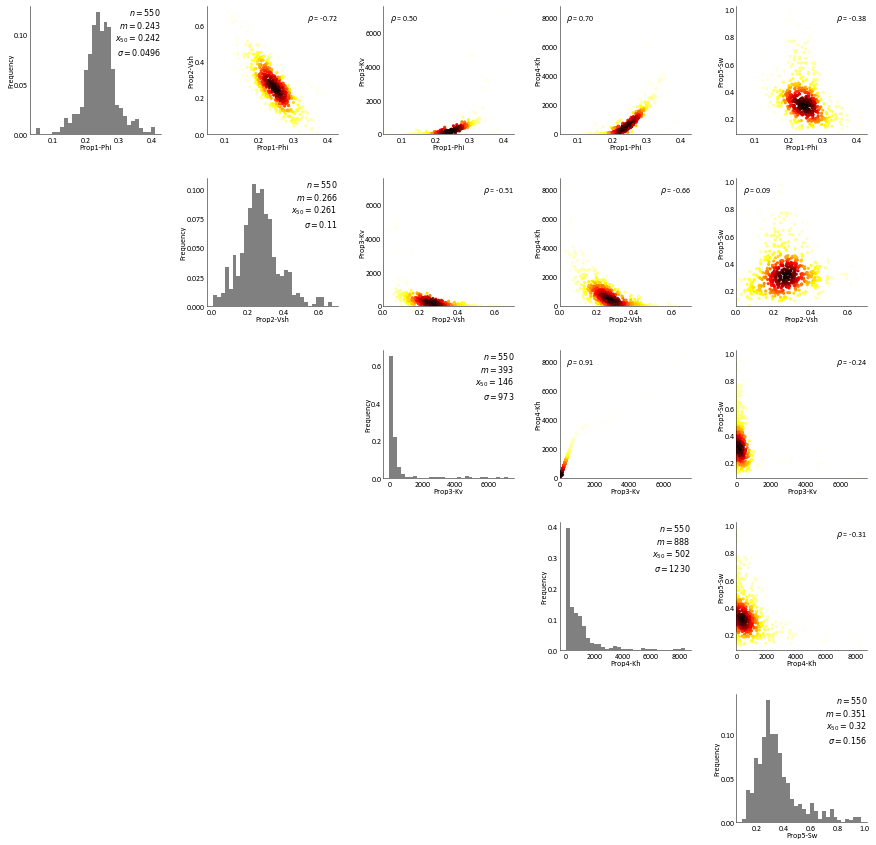
RockType 2¶
[17]:
dat = Well_DF.data[Well_DF.data['RockType']==2]
# setup some custom styles for the output
pltstyle = 'ccgpaper'
cust_style = None
# Upper Triangle: KDE plot
def iter_upper(axes):
for ax, (varx, vary) in zip(axes, utrivars):
gs.kdeplot(dat[varx], dat[vary], ax=ax, density=False, rotateticks=(False, False), cmap="hot_r",
cbar=False, contour=True, lw=0.5, pltstyle=pltstyle, cust_style=cust_style, s=10,
threshold=0.1, shade=False)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Diagonal: Histograms
def iter_diagonal(axes):
for ax, var in zip(axes, variables):
gs.histplt(dat[var], ax=ax, color="grey", pltstyle=pltstyle, cust_style=cust_style,
xlabel=var)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Setup the imagegrid using symmetric, the three functions, and the `unequal_aspects` keyword
# which allows the histograms to be plotted along side the bivariate plots without strange aspect
# issues
fig = gs.imagegrid(upperfunc=iter_upper, diagfunc=iter_diagonal, nvar=len(variables),
symmetric=True, unequal_aspects=True, axes_pad=(0.35, 0.35),
ntickbins=(6,5), pltstyle=pltstyle, cust_style=cust_style, figsize=(15, 15))
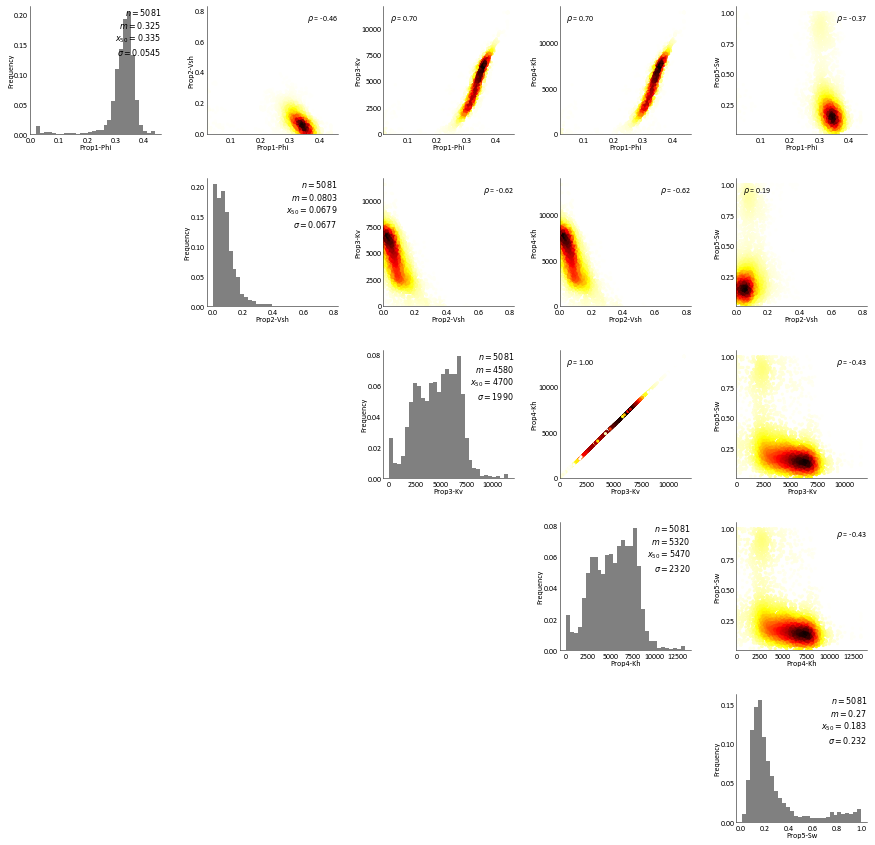
RockType 3¶
[18]:
dat = Well_DF.data[Well_DF.data['RockType']==3]
# setup some custom styles for the output
pltstyle = 'ccgpaper'
cust_style = None
# Upper Triangle: KDE plot
def iter_upper(axes):
for ax, (varx, vary) in zip(axes, utrivars):
gs.kdeplot(dat[varx], dat[vary], ax=ax, density=False, rotateticks=(False, False), cmap="hot_r",
cbar=False, contour=True, lw=0.5, pltstyle=pltstyle, cust_style=cust_style, s=10,
threshold=0.1, shade=False)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Diagonal: Histograms
def iter_diagonal(axes):
for ax, var in zip(axes, variables):
gs.histplt(dat[var], ax=ax, color="grey", pltstyle=pltstyle, cust_style=cust_style,
xlabel=var)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Setup the imagegrid using symmetric, the three functions, and the `unequal_aspects` keyword
# which allows the histograms to be plotted along side the bivariate plots without strange aspect
# issues
fig = gs.imagegrid(upperfunc=iter_upper, diagfunc=iter_diagonal, nvar=len(variables),
symmetric=True, unequal_aspects=True, axes_pad=(0.35, 0.35),
ntickbins=(6,5), pltstyle=pltstyle, cust_style=cust_style, figsize=(15, 15))
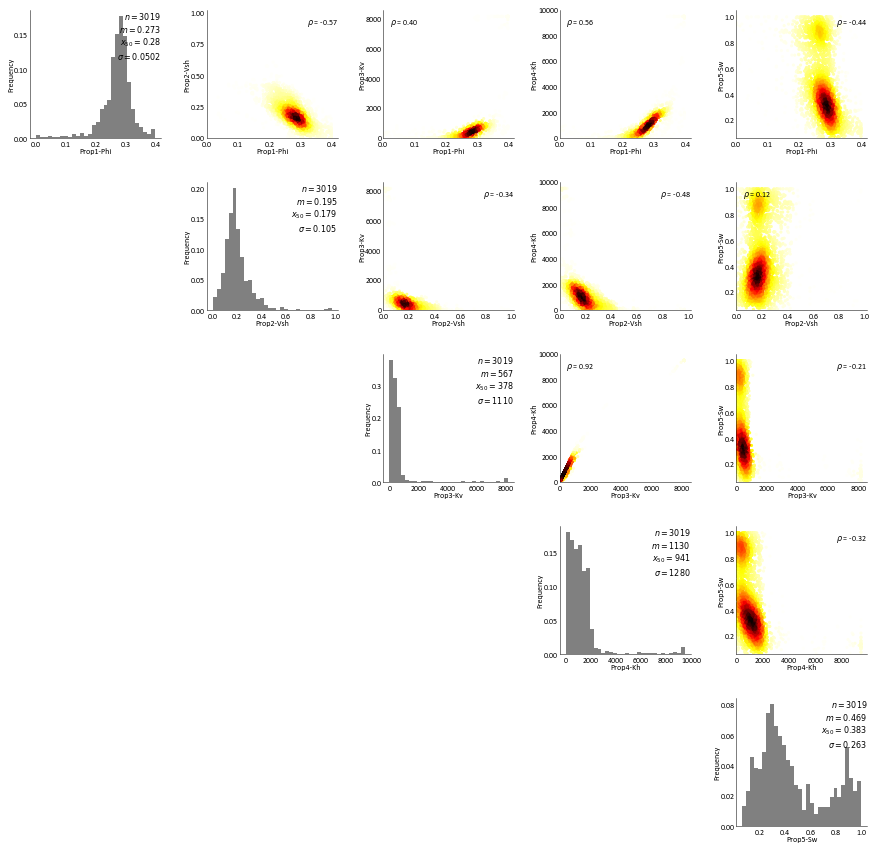
RockType 4¶
[19]:
dat = Well_DF.data[Well_DF.data['RockType']==4]
# setup some custom styles for the output
pltstyle = 'ccgpaper'
cust_style = None
# Upper Triangle: KDE plot
def iter_upper(axes):
for ax, (varx, vary) in zip(axes, utrivars):
gs.kdeplot(dat[varx], dat[vary], ax=ax, density=False, rotateticks=(False, False), cmap="hot_r",
cbar=False, contour=True, lw=0.5, pltstyle=pltstyle, cust_style=cust_style, s=10,
threshold=0.1, shade=False)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Diagonal: Histograms
def iter_diagonal(axes):
for ax, var in zip(axes, variables):
gs.histplt(dat[var], ax=ax, color="grey", pltstyle=pltstyle, cust_style=cust_style,
xlabel=var)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Setup the imagegrid using symmetric, the three functions, and the `unequal_aspects` keyword
# which allows the histograms to be plotted along side the bivariate plots without strange aspect
# issues
fig = gs.imagegrid(upperfunc=iter_upper, diagfunc=iter_diagonal, nvar=len(variables),
symmetric=True, unequal_aspects=True, axes_pad=(0.35, 0.35),
ntickbins=(6,5), pltstyle=pltstyle, cust_style=cust_style, figsize=(15, 15))
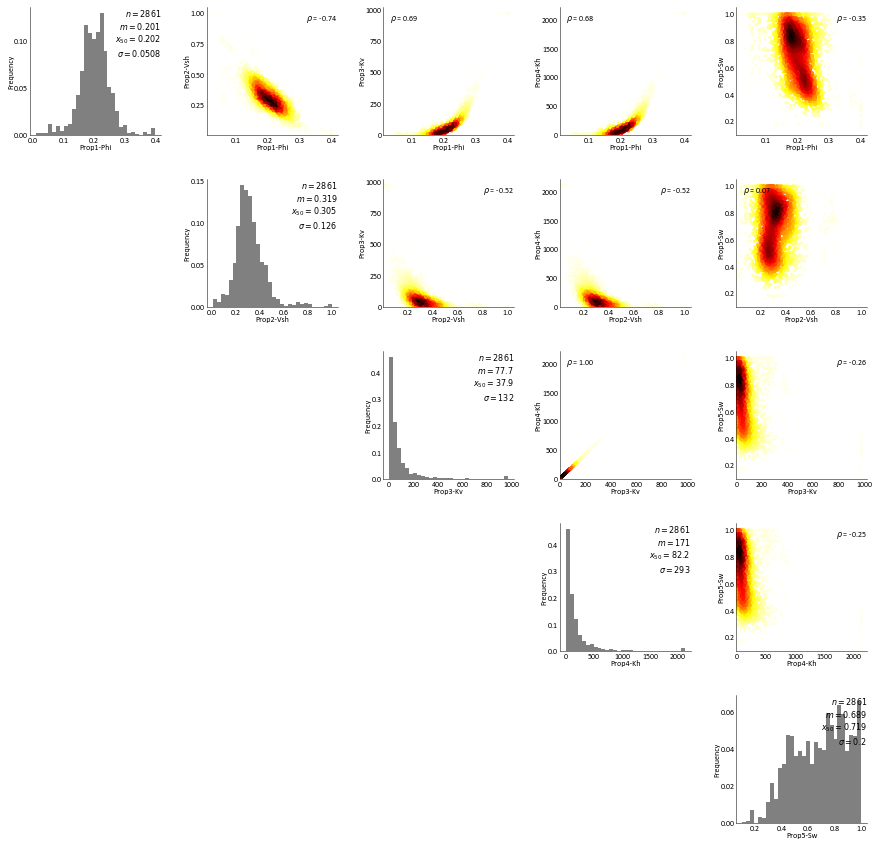
RockType 5¶
[20]:
dat = Well_DF.data[Well_DF.data['RockType']==5]
# setup some custom styles for the output
pltstyle = 'ccgpaper'
cust_style = None
# Upper Triangle: KDE plot
def iter_upper(axes):
for ax, (varx, vary) in zip(axes, utrivars):
gs.kdeplot(dat[varx], dat[vary], ax=ax, density=False, rotateticks=(False, False), cmap="hot_r",
cbar=False, contour=True, lw=0.5, pltstyle=pltstyle, cust_style=cust_style, s=10,
threshold=0.1, shade=False)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Diagonal: Histograms
def iter_diagonal(axes):
for ax, var in zip(axes, variables):
gs.histplt(dat[var], ax=ax, color="grey", pltstyle=pltstyle, cust_style=cust_style,
xlabel=var)
# tighten the label spacing
ax.yaxis.labelpad = 0
ax.xaxis.labelpad = 0
ax.tick_params(axis='both', which='major', pad=0)
yield ax
# Setup the imagegrid using symmetric, the three functions, and the `unequal_aspects` keyword
# which allows the histograms to be plotted along side the bivariate plots without strange aspect
# issues
fig = gs.imagegrid(upperfunc=iter_upper, diagfunc=iter_diagonal, nvar=len(variables),
symmetric=True, unequal_aspects=True, axes_pad=(0.35, 0.35),
ntickbins=(6,5), pltstyle=pltstyle, cust_style=cust_style, figsize=(15, 15))